|
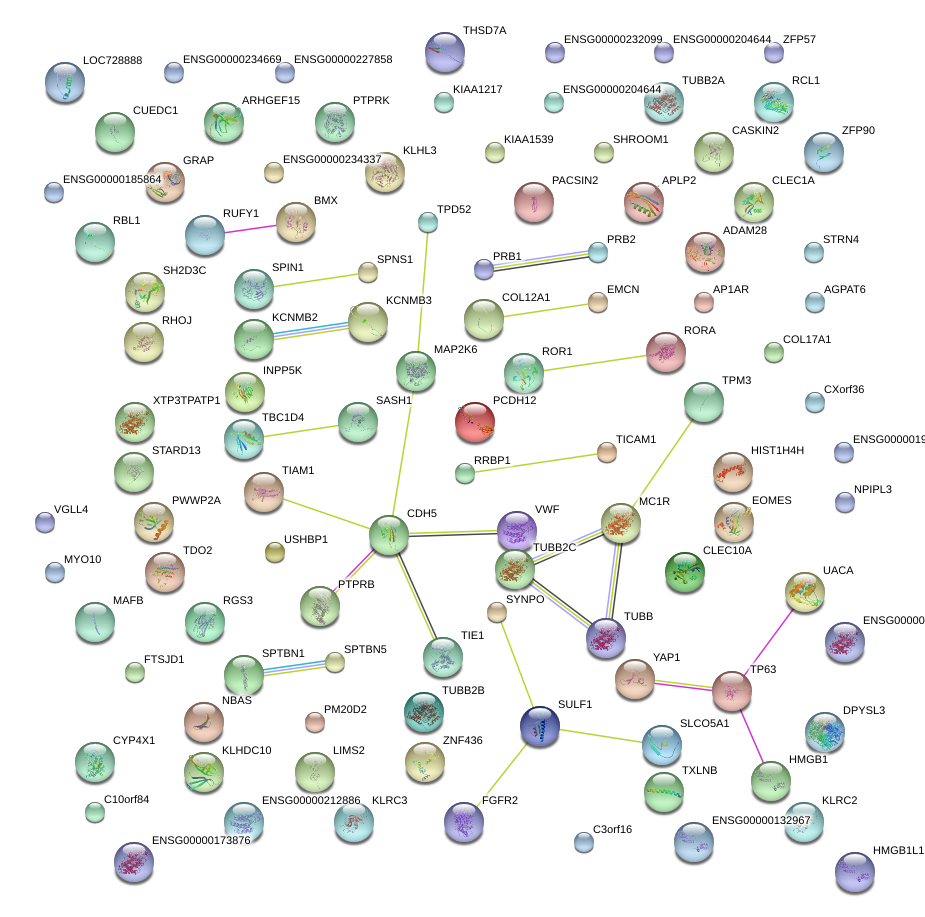
chr12_-_6103945
|
1.581
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr11_+_129496792
|
1.175
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr1_-_197173159
|
1.022
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr7_-_11838260
|
1.017
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr14_-_89490780
|
0.996
|
NM_145231
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chr16_+_22432344
|
0.991
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chrX_+_15428820
|
0.925
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr16_-_21344158
|
0.913
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr16_-_69876262
|
0.889
|
|
FTSJD1
|
FtsJ methyltransferase domain containing 1
|
|
chr6_-_139654807
|
0.863
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chrX_-_44945025
|
0.818
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr17_-_71017509
|
0.815
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr5_+_149960820
|
0.758
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr2_-_128138435
|
0.748
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr19_-_17236522
|
0.734
|
NM_031941
|
USHBP1
|
Usher syndrome 1C binding protein 1
|
|
chr5_-_141318810
|
0.731
|
NM_016580
|
PCDH12
|
protocadherin 12
|
|
chr10_-_123346148
|
0.719
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr17_-_1359745
|
0.715
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr8_-_70908130
|
0.714
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr3_+_182900081
|
0.693
|
|
SOX2OT
|
SOX2 overlapping transcript (non-protein coding)
|
|
chr20_-_38751289
|
0.675
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr6_-_29752909
|
0.661
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr7_+_129497808
|
0.658
|
|
KLHDC10
|
kelch domain containing 10
|
|
chr9_-_35101592
|
0.618
|
|
KIAA1539
|
KIAA1539
|
|
chr1_+_43539239
|
0.614
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr17_+_8154280
|
0.614
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr17_+_65009986
|
0.613
|
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr8_+_24207524
|
0.612
|
NM_014265
NM_021777
|
ADAM28
|
ADAM metallopeptidase domain 28
|
|
chr3_-_11598829
|
0.612
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr14_+_62740878
|
0.608
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr16_+_64958060
|
0.606
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr17_-_18890963
|
0.602
|
NM_006613
|
GRAP
|
GRB2-related adaptor protein
|
|
chr5_-_137099602
|
0.600
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr9_+_4782974
|
0.597
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr3_-_150993299
|
0.590
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr6_+_148705421
|
0.581
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr10_+_24538094
|
0.579
|
|
KIAA1217
|
KIAA1217
|
|
chr5_-_146813343
|
0.578
|
NM_001387
|
DPYSL3
|
dihydropyrimidinase-like 3
|
|
chr3_+_179736917
|
0.571
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr6_-_75972473
|
0.566
|
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr5_-_159478967
|
0.562
|
NM_001130864
NM_052927
|
PWWP2A
|
PWWP domain containing 2A
|
|
chr2_+_54536780
|
0.559
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr16_+_64958025
|
0.558
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr8_+_70541631
|
0.552
|
|
SULF1
|
sulfatase 1
|
|
chr9_+_115303527
|
0.549
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr9_-_35101534
|
0.548
|
|
KIAA1539
|
KIAA1539
|
|
chr2_-_56266408
|
0.546
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr9_-_129557373
|
0.546
|
NM_001142534
NM_001142533
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr15_-_68781662
|
0.543
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr15_-_39973566
|
0.543
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr9_+_4782833
|
0.541
|
NM_005772
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr5_+_178919298
|
0.541
|
NM_001040451
NM_001040452
|
RUFY1
|
RUN and FYVE domain containing 1
|
|
chr16_+_67131161
|
0.538
|
NM_133458
|
ZFP90
|
zinc finger protein 90 homolog (mouse)
|
|
chr12_-_10142779
|
0.536
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr3_+_190990142
|
0.532
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr6_-_75972286
|
0.532
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr8_-_81155564
|
0.526
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr6_-_128883125
|
0.520
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr9_+_4782876
|
0.512
|
|
RCL1
|
RNA terminal phosphate cyclase-like 1
|
|
chr22_-_41672990
|
0.512
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr1_-_152431175
|
0.512
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr9_+_90193807
|
0.511
|
|
SPIN1
|
spindlin 1
|
|
chr12_-_11399776
|
0.510
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chr13_-_32658215
|
0.509
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr16_+_64958086
|
0.505
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr10_+_24537725
|
0.505
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr17_-_53335748
|
0.504
|
NM_017949
|
CUEDC1
|
CUE domain containing 1
|
|
chr10_-_91451102
|
0.504
|
|
NBAS
|
neuroblastoma amplified sequence
|
|
chr1_+_64012154
|
0.501
|
NM_001083592
NM_005012
|
ROR1
|
receptor tyrosine kinase-like orphan receptor 1
|
|
chr10_-_105835609
|
0.500
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr13_-_74799898
|
0.499
|
|
TBC1D4
|
TBC1 domain family, member 4
|
|
chr3_-_14964861
|
0.497
|
|
LOC100505673
|
hypothetical LOC100505673
|
|
chr10_-_120091795
|
0.496
|
NM_001134672
NM_022063
|
C10orf84
|
chromosome 10 open reading frame 84
|
|
chr20_-_17610828
|
0.492
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr12_-_120725210
|
0.489
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr8_+_121892719
|
0.489
|
|
|
|
|
chr5_+_95093267
|
0.486
|
|
|
|
|
chr4_-_101658094
|
0.484
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr8_+_70541412
|
0.476
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr5_-_137099361
|
0.474
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr21_-_31853160
|
0.473
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr3_-_27739201
|
0.467
|
|
EOMES
|
eomesodermin
|
|
chr1_-_23568854
|
0.464
|
NM_001077195
|
ZNF436
|
zinc finger protein 436
|
|
chr11_+_101488380
|
0.454
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr13_-_29938029
|
0.453
|
NM_002128
|
HMGB1
|
high-mobility group box 1
|
|
chr12_-_69289889
|
0.451
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr1_+_47261826
|
0.447
|
NM_178033
|
CYP4X1
|
cytochrome P450, family 4, subfamily X, polypeptide 1
|
|
chr4_+_113372287
|
0.447
|
NM_001128426
NM_018569
|
AP1AR
|
adaptor-related protein complex 1 associated regulatory protein
|
|
chr5_-_16989371
|
0.444
|
NM_012334
|
MYO10
|
myosin X
|
|
chr12_-_10464308
|
0.441
|
NM_002261
NM_007333
|
KLRC3
|
killer cell lectin-like receptor subfamily C, member 3
|
|
chr5_-_132189762
|
0.441
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr10_-_105835525
|
0.440
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr8_-_70907957
|
0.438
|
NM_001146009
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr6_+_30796064
|
0.432
|
|
TUBB
|
tubulin, beta
|
|
chr3_-_189354510
|
0.432
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr8_-_38444398
|
0.431
|
NM_001174065
NM_001174066
NM_001174067
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr12_-_11439752
|
0.426
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr11_-_46824364
|
0.426
|
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr2_+_228386800
|
0.423
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr1_+_176577228
|
0.422
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr13_+_51927615
|
0.421
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr2_+_37425240
|
0.419
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr12_-_10173947
|
0.418
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr10_-_13789628
|
0.418
|
|
FRMD4A
|
FERM domain containing 4A
|
|
chr1_+_149395728
|
0.415
|
NM_024575
|
TNFAIP8L2
|
tumor necrosis factor, alpha-induced protein 8-like 2
|
|
chr17_+_44325127
|
0.411
|
NM_001002027
NM_005175
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9)
|
|
chr14_-_63830830
|
0.408
|
NM_001040275
NM_001437
|
ESR2
|
estrogen receptor 2 (ER beta)
|
|
chr5_-_92942680
|
0.408
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr14_+_58725104
|
0.404
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr17_-_36996611
|
0.400
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr1_+_183970093
|
0.399
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr10_+_115300579
|
0.399
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr10_+_104253706
|
0.397
|
NM_001178133
NM_016169
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr4_-_186969202
|
0.395
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_-_121892877
|
0.393
|
|
SNTB1
|
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1)
|
|
chr1_+_200449905
|
0.391
|
NM_001017404
|
LGR6
|
leucine-rich repeat containing G protein-coupled receptor 6
|
|
chr12_-_57600564
|
0.390
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr12_+_14429499
|
0.389
|
|
ATF7IP
|
activating transcription factor 7 interacting protein
|
|
chr7_+_134114912
|
0.389
|
|
CALD1
|
caldesmon 1
|
|
chr3_-_101077605
|
0.388
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr5_+_50714996
|
0.388
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr7_-_27206249
|
0.387
|
NM_000522
|
HOXA13
|
homeobox A13
|
|
chr7_-_27186400
|
0.387
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr1_+_151270301
|
0.384
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr3_+_44571600
|
0.382
|
NM_018651
NM_025169
|
ZNF167
|
zinc finger protein 167
|
|
chr10_-_103864579
|
0.379
|
|
LDB1
|
LIM domain binding 1
|
|
chr1_+_34997928
|
0.375
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr20_-_17610704
|
0.374
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr4_-_175441192
|
0.373
|
|
FBXO8
|
F-box protein 8
|
|
chr6_+_21774624
|
0.369
|
|
FLJ22536
|
hypothetical locus LOC401237
|
|
chr4_-_186969041
|
0.368
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_-_64087806
|
0.368
|
NM_001143940
NM_016571
|
LGSN
|
lengsin, lens protein with glutamine synthetase domain
|
|
chr11_-_46824416
|
0.366
|
NM_001008938
NM_014756
|
CKAP5
|
cytoskeleton associated protein 5
|
|
chr7_-_13997389
|
0.366
|
NM_001163149
|
ETV1
|
ets variant 1
|
|
chr1_-_148124738
|
0.364
|
NM_003528
|
HIST2H2BE
|
histone cluster 2, H2be
|
|
chr15_-_70277163
|
0.363
|
NM_001012642
|
GRAMD2
|
GRAM domain containing 2
|
|
chr7_+_134114957
|
0.362
|
|
CALD1
|
caldesmon 1
|
|
chr1_+_159495140
|
0.359
|
NM_001102566
|
PCP4L1
|
Purkinje cell protein 4 like 1
|
|
chr5_+_60276695
|
0.358
|
NM_174889
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2
|
|
chr8_-_82557956
|
0.356
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr18_+_3437607
|
0.356
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_-_24606616
|
0.354
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr4_-_149582754
|
0.353
|
|
NR3C2
|
nuclear receptor subfamily 3, group C, member 2
|
|
chr9_+_90193058
|
0.352
|
NM_006717
|
SPIN1
|
spindlin 1
|
|
chr5_-_50714836
|
0.352
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr4_-_151990061
|
0.351
|
|
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing
|
|
chr17_-_59818033
|
0.350
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_+_83627422
|
0.348
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr14_-_72563497
|
0.345
|
NM_021260
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr2_-_207739815
|
0.345
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr7_-_41709191
|
0.345
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr5_-_16989290
|
0.342
|
|
MYO10
|
myosin X
|
|
chr15_-_57013092
|
0.341
|
NM_001013843
NM_024755
|
SLTM
|
SAFB-like, transcription modulator
|
|
chr20_-_17459973
|
0.340
|
NM_001195
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr5_+_159781454
|
0.339
|
|
PTTG1
|
pituitary tumor-transforming 1
|
|
chr14_+_101297887
|
0.336
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr7_-_13997574
|
0.336
|
NM_001163148
|
ETV1
|
ets variant 1
|
|
chr11_+_116608447
|
0.335
|
NM_207343
NM_001077239
|
RNF214
|
ring finger protein 214
|
|
chr12_-_47362254
|
0.334
|
NM_017822
|
C12orf41
|
chromosome 12 open reading frame 41
|
|
chr19_-_63205948
|
0.334
|
|
ZNF606
|
zinc finger protein 606
|
|
chr12_-_97812633
|
0.332
|
|
ANKS1B
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr14_-_72523404
|
0.330
|
NM_178441
|
ZFYVE1
|
zinc finger, FYVE domain containing 1
|
|
chr6_+_27208763
|
0.329
|
NM_021064
|
HIST1H2AG
|
histone cluster 1, H2ag
|
|
chr9_+_123501186
|
0.328
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr6_-_116681863
|
0.328
|
NM_021648
|
TSPYL4
|
TSPY-like 4
|
|
chr14_-_54948290
|
0.328
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr14_+_90650466
|
0.327
|
|
C14orf159
|
chromosome 14 open reading frame 159
|
|
chr13_-_32678142
|
0.325
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr4_+_87070449
|
0.325
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr3_-_194118627
|
0.322
|
NM_178496
|
MB21D2
|
Mab-21 domain containing 2
|
|
chr2_-_96769502
|
0.322
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr3_+_115099007
|
0.322
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chr6_+_64289585
|
0.321
|
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr3_+_113200696
|
0.320
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr10_-_103864651
|
0.316
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr3_+_113200275
|
0.316
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr3_+_189354167
|
0.315
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr10_-_94040779
|
0.315
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr4_+_57066131
|
0.314
|
NM_206919
|
ARL9
|
ADP-ribosylation factor-like 9
|
|
chr10_+_91451313
|
0.307
|
NM_016195
|
KIF20B
|
kinesin family member 20B
|
|
chr11_-_128567302
|
0.307
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr4_+_175441402
|
0.307
|
NM_001040157
|
KIAA1712
|
KIAA1712
|
|
chr20_+_10363949
|
0.306
|
NM_001009608
|
C20orf94
|
chromosome 20 open reading frame 94
|
|
chr1_+_42701460
|
0.306
|
|
CCDC30
|
coiled-coil domain containing 30
|
|
chr14_-_94306104
|
0.306
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr6_-_72187168
|
0.305
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr14_+_36196523
|
0.303
|
NM_006194
|
PAX9
|
paired box 9
|
|
chr6_+_26153590
|
0.302
|
NM_003531
|
HIST1H3C
|
histone cluster 1, H3c
|
|
chr4_-_106849252
|
0.301
|
NM_001142471
NM_020395
|
INTS12
|
integrator complex subunit 12
|
|
chr1_+_246635918
|
0.300
|
NM_030904
|
OR2T1
|
olfactory receptor, family 2, subfamily T, member 1
|
|
chr7_-_27108826
|
0.298
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr5_+_15553288
|
0.298
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr9_-_98104206
|
0.298
|
NM_000197
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3
|
|
chr5_+_57914658
|
0.297
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr20_-_13567529
|
0.297
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr10_+_32896656
|
0.296
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr1_-_24311212
|
0.296
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr4_+_41235077
|
0.295
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_-_100088760
|
0.295
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr13_-_95094920
|
0.294
|
NM_014934
NM_198968
|
DZIP1
|
DAZ interacting protein 1
|